

CASTpFold provides detailed quantitative analysis of geometric and topological properties of protein structures, including pockets (AF2 ID: A0A2N0QT79 Fig 1B), interior cavities (no mouth), and cross channels(PDB ID: 5BUN Fig 1C).

In the "Pocket Similarity" panel, CASTpFold measures the similarity among the database of surface pockets and protein-protein interface (PPI) pockets. An exmaple of surface pocket is shown in Fig 2A (AF2 ID: A0A0C3Q0M4), and a PPI pocket is shown in Fig 2B (PDB ID: 6M0J).
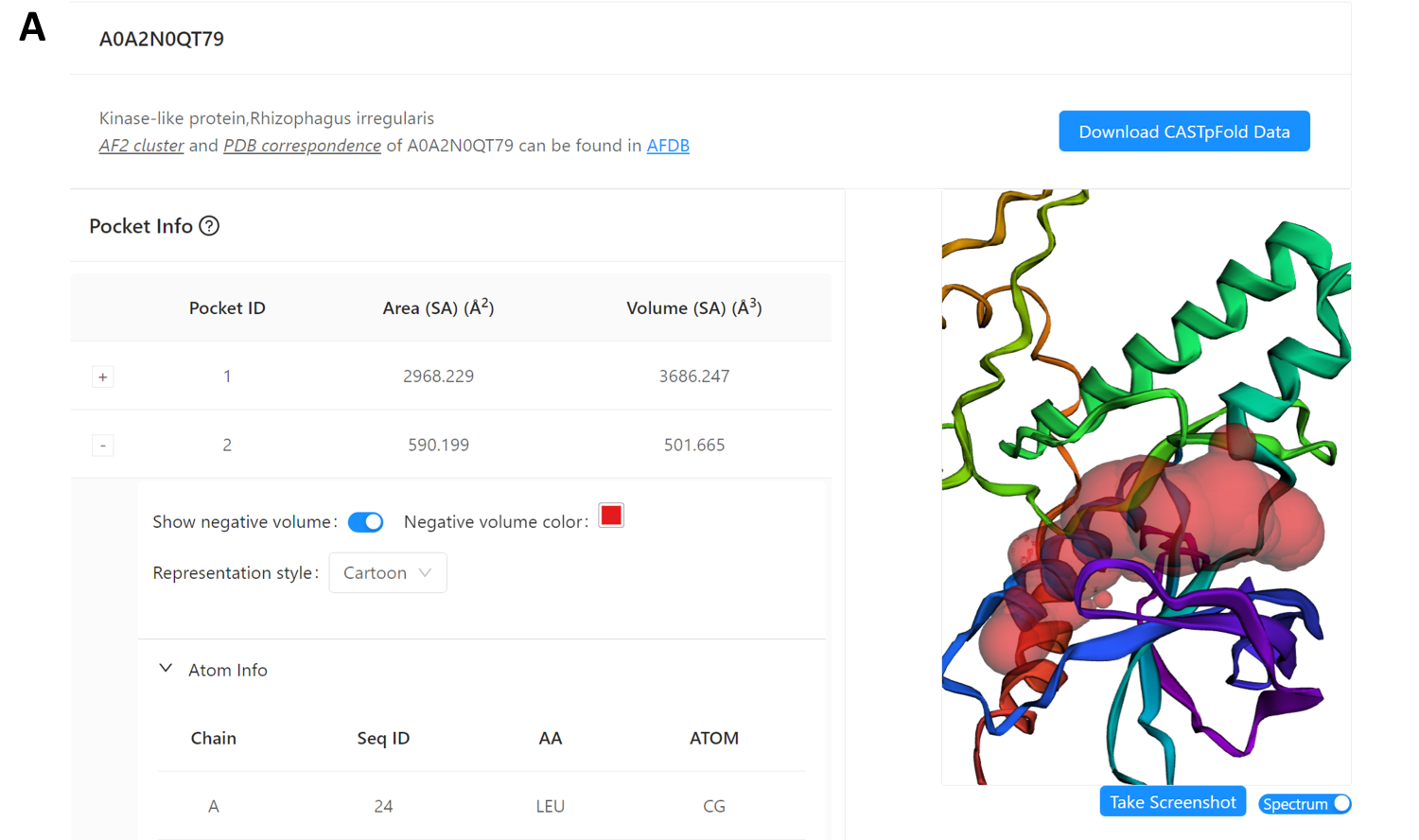
In Fig 3, the “Basic Info” panel for a AF2 predicted protein (AF2 ID: A0A2N0QT79) displays information from the UniProt database. The “Pocket Info” panel depicts the selected pocket (Pocket ID: 2), which is rendered in cartoon style in the “Structure viewer” panel. Additionally, an expandable “Atom Info” panel reveals the atomic details of the selected pocket. Users can identify the residue positions within the structure in the viewer by clicking the corresponding row.

Fig 4 shows the “Sequence Info” panel for the structure (AF2 ID: A0A2N0QT79), including pocket residue with atom information, which is visualized as cyan spheres in the viewer.
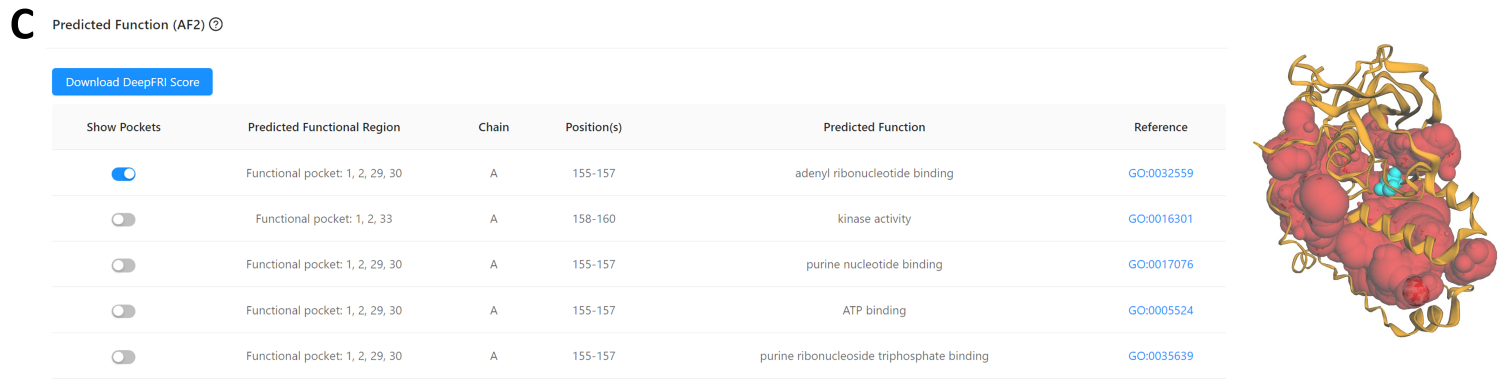
Users exploring the AF2-predicted structure (AF2 ID: A0A2N0QT79) can find a “Predicted Function” panel (Fig 5), which lists the predicted function of the pocket, including the associated GO terms or EC numbers, functionally relevant positions, and links to details of potential functions. Users can download the corresponding DeepFRI prediction score using the "Download DeepFRI Score" button.

Fig 6 shows a pocket from an the structure (AF2 ID: A0A2N0QT79, Pocket ID: 2) that has a total of 93 pockets across both PDB and AF2 structures exhibiting similarity to the queried pocket. One of these is from the PDB structure (PDB ID: 7NQQ, Pocket ID: 1), which is visualized in the “Similar pocket viewer”.

In Fig 7, the “Basic Info” panel for a PDB structure (PDB ID: 6M0J) displays information from the UniProt database. The “Pocket Info” panel depicts the selected pocket (Pocket ID: 1), which is rendered in cartoon style in the “Structure viewer” panel.
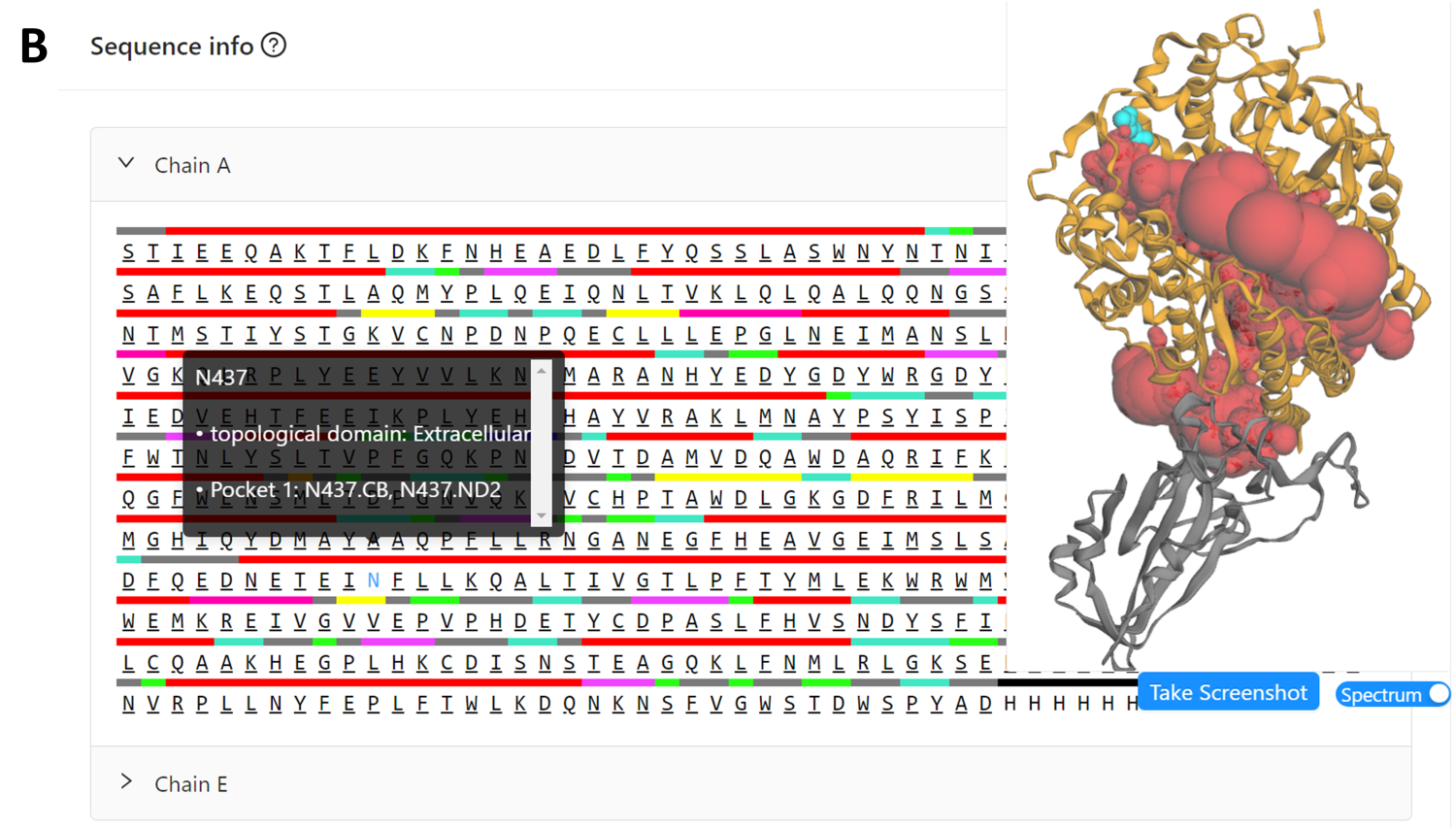
Fig 8 shows the “Sequence Info” panel for the PDB structure (PDB ID: 6M0J), including annotation and pocket residue with atom information, which is visualized as cyan spheres in the viewer.

In Fig 9, an “Annotation” panel is available for PDB structures, providing annotations extracted from the UniProt database. PDB annotated residues can be visualized together with their respective pockets in a combined view. Here, pockets 1, 28, and 45 are visualized alongside the annotated residues spanning from 30 to 41.
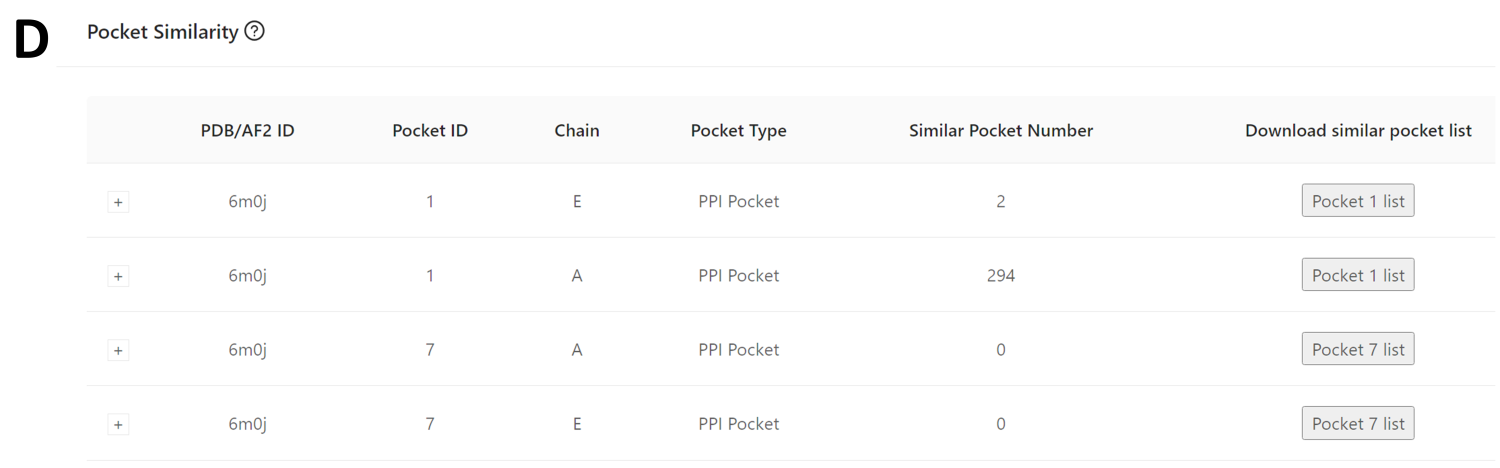
Fig 10 presents four pockets containing at least 14 residues identified within the PDB structure (PDB ID: 6M0J). Among these, two pockets have a nonzero number of similar members: Pocket 1 on chain A boasts 294 similar members, and chain E includes 2 members.

The 'PDB counterpart' panel facilitates the comparison between PDB entries and their AF2 counterparts. Users can compare PDB subunits with their corresponding AF2 structures by clicking the relevant link. Figure 11 illustrates that chain A of the PDB structure (PDB ID: 6M0J) corresponds to UniProt ID: Q98YF1, which is mapped to the AF2 predicted representative structure (AF2 ID: (PDB ID: A0A7R9PMA8)). Conversely, chain E corresponds to UniProt ID: P0DTC2, which cannot be mapped to any AF2 predicted representative structure.